-
Notifications
You must be signed in to change notification settings - Fork 3
Usage examples
- Prerequisites - installation and sample data
- Transforming images - code for rendering and transforming images
- Transforming neuron skeletons - code for transforming neurons in swc format
- Converting transforms
-
CMTK example - converting a h5 transform to CMTK-compatible format and applying it with CMTK's
reformatx. - Processing swc's - process / edit swc neuron files
- Install jgo
- Clone and install this repository:
git clone https://github.com/saalfeldlab/template-building.git
cd template-building
git checkout v0.1.0
mvn clean compile install
Some of these examples use sample data that can be downloaded from figshare.
Include are:
- A downsampled version of the JRC 2018 female template.
- A downsampled version of the FCWB template.
The neuron skeleton included here comes from the FlyCircuit database and accessed through Greg Jefferis' flycircuit R package
We recommend using the transformImage shell script for transforming images.
Usage: transformImage [-v] [--interpolation=<interpolation>] [-j=<nThreads>]
[-s=<outputSize>] -i=<inputFiles> [-i=<inputFiles>]...
-o=<outputFiles> [-o=<outputFiles>]...
[-r=<outputResolution>[,<outputResolution>...]]...
[-t=<transformFiles>]...
--interpolation=<interpolation>
Interpolation {LINEAR, NEAREST, LANCZOS}
-i, --input=<inputFiles> Image file to transform
-j, --nThreads=<nThreads> Number of rendering threads (default=1)
-o, --output=<outputFiles> Output file for transformed image
-r, --output-resolution=<outputResolution>[,<outputResolution>...]
The resolution at which to write the output
-s, --outputImageSize=<outputSize>
Size of image output in pixels
-t, --transform=<transformFiles>
Transformation file.
-v, --version Prints version information and exits.
Uses this sample data.
transformImage -i FCWB_small.nrrd \
-o fcwb-to-jrc18f.nrrd \
-f JRC2018_FEMALE_small.nrrd \
-t JRC2018F_FCWB_small.h5 \
-j 4
Summary of arguments:
-
-i FCWB_small.nrrd- the input image we to transform is the
FCWBtemplate
- the input image we to transform is the
-
-o fcwb-to-jrc18f.nrrd- give the name of the output image
-
-f JRC2018_FEMALE_small.nrrd- the field-of-view and image resolutions of the output image should be that of the
JRC2018Ftemplate
- the field-of-view and image resolutions of the output image should be that of the
-
-t JRC2018F_FCWB_small.h5- use the transform mapping
JRC2018FtoFCWBsee here for details on this nomenclature and directions of transforms.
- use the transform mapping
-
-j 4- Use four threads for parallel rendering
Uses this sample data.
transformImage -i JRC2018_FEMALE_small.nrrd \
-o jrc18f-to-fcwb.nrrd \
-f FCWB_small.nrrd \
-t JRC2018F_FCWB_small.h5??i \
-j 4
Summary of arguments:
-
-i JRC2018_FEMALE_small.nrrd- the input image we to transform is the
FCWBtemplate
- the input image we to transform is the
-
-o jrc18f-to-fcwb.nrrd- give the name of the output image
-
-f FCWB_small.nrrd- the field-of-view and image resolutions of the output image should be that of the
JRC2018Ftemplate
- the field-of-view and image resolutions of the output image should be that of the
-
-t JRC2018F_FCWB_small.h5??i -
-j 4- Use four threads for parallel rendering
These examples show how to transform neuronal skeletons stored as swc files. We recommend using the transformSwc shell script or the transform_swc fiji script.
You can see a summary of the options by running
$ transformSwc --help
Usage: <main class> [-hv] [--dry-run] [--exclude=<excludeMatcher>]
[--include=<includeMatcher>] [--suffix=<suffix>]
[-d=<skeletonDirectory>] [-o=<outputSkeletonPaths>]...
[-s=<skeletonPaths>]... [-t=<transforms>]...
--dry-run Prints files to work on, but does nothing.
--exclude=<excludeMatcher>
Exclusion pattern: Files matching the given regular
expression will be ignored. Only relevant when pass a
directory
--include=<includeMatcher>
Matching pattern: Files not matching the given regular
expression will be ignored. Only relevant when passing
a directory
--suffix=<suffix> Suffix applied to output files when passing a directory.
-d, --directory=<skeletonDirectory>
Directory containing skeletons
-h, --help Print this help message
-o, --output=<outputSkeletonPaths>
Output skeleton file names. Must have one output for every
input skeleton, and pass outputfile paths in the same
order as the input sketons.
-s, --skeleton=<skeletonPaths>
Input skeletons. Can pass multiple skeletons
-t, --transform=<transforms>
List of transforms. Every transform that is passed will
be applied to the skeletons in the order they are
passed. -t <transform file path> applies the inverse of
the given transform if possible.
-v, --version Prints version information and exits.
Transform an skeleton and write it to an output file
transformSwc -s <swc skeleton file> -o <output file> -t <transform file>
Transform multiple skeletons with the same transforms:
transformSwc \
-s <swc skeleton file 1> -o <output file 1> \
-s <swc skeleton file 2> -o <output file 2> \
-t <transform file>
Transform a skeleton with a sequence of transforms (some inverted). In this example the inverse of the third transform is applied:
transformSwc \
-s <swc skeleton file> -o <output file> \
-t <transform file 1> -t <transform file 2> -t inverse <transform file 3>
transformSwc can operate on all files in a directory
transformSwc -d <directory> -t <transform>
applies the transform to all swc files in directory. Use of the -d flag overrides any skeletons passed with -s flags. In this case, the suffix _transformed is appended to any processed files. The suffix can be changed using the ``--suffix` flag:
transformSwc -d <directory> -t <transform> --suffix _hippo
If you want to process some, not all, of the files in a directory, the --include`` and --exclude` flags let you pass a regular expression for files to include and exclude, respectively. Note inclusions are are processed before exclusions.
Example, to process files in a directory starting with an "a":
transformSwc -d <directory> -t <transform> --include '^a.*'
Example, to exclude files with a number anywhere in the file name:
transformSwc -d <directory> -t <transform> --exclude '.*[0-9].*'
Uses this sample data.
transformNeuron \
-s GadMARCM-F000122_seg001_03.swc \
-o GadMARCM_xfm-to-jrc18f.swc \
-t JRC2018F_FCWB_small.h5??i
Summary of arguments:
-
s GadMARCM-F000122_seg001_03.swc- the input skeleton to transform
-
-o GadMARCM_xfm-to-jrc18f.swc- the name of the output skeleton
-
-t JRC2018F_FCWB_small.h5??i- use the inverse of the
JRC2018F_FCWBbecause the inverse maps points fromFCWBtoJRC2018Fsee here for details on this nomenclature and directions of transformations.
- use the inverse of the
| FCWB space | JRC2018F space |
|---|---|
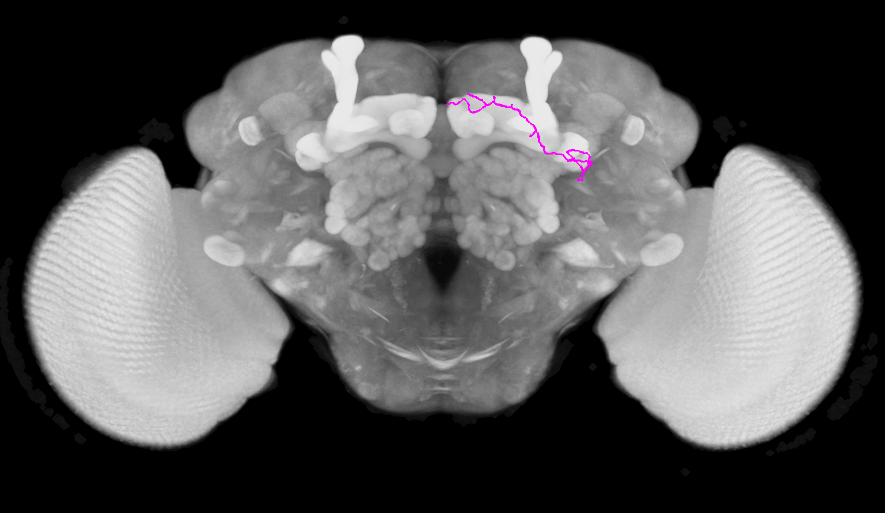 |
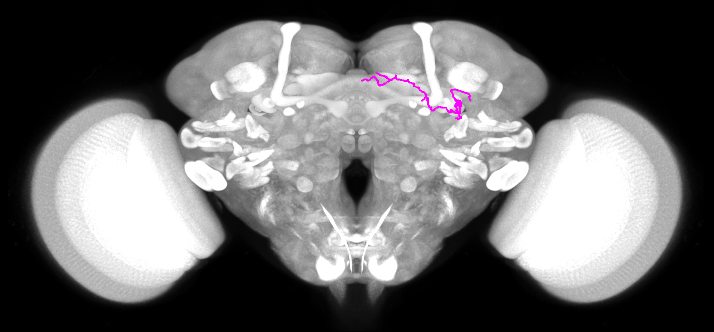 |
Uses this sample data.
convertAffine JRC2018F_FCWB_small.h5 JRC2018_FEMALE_small.xform
# convert forward deformation field
convertDfield JRC2018F_FCWB_small.h5 JRC2018F_FCWB_small_fwd.nrrd
# convert inverse deformation field
convertDfield JRC2018F_FCWB_small.h5??i JRC2018F_FCWB_small_inv.nrrd
# Use cmtk to apply transform to FCWB
reformatx --floating FCWB_small.nrrd \
-o cmtk_fcwb-to-jrc18f.nrrd \
JRC2018_FEMALE_small.nrrd \
JRC2018F_FCWB_small_fwd.nrrd JRC2018_FEMALE_small.xform
# Use cmtk to apply transform to JRC2018F
reformatx --floating JRC2018_FEMALE_small.nrrd \
-o cmtk_jrc18f-to-fcwb.nrrd \
FCWB_small.nrrd \
-i JRC2018_FEMALE_small.xform JRC2018F_FCWB_small_inv.nrrd
-
convertAffine-
xformis cmtk's extension for transformations
-
It may sometimes be useful to concatenate transformations to a single file. Deformation fields, being the most general representation, is a natural choice. The transform2Dfield implements this.
Usage: <main class> [-f=<referenceImagePath>] -o=<outputFile> [-q=<nThreads>]
[-s=<outputSize>] [-r=<outputResolution>[,
<outputResolution>...]]... [-t=<transformFiles>]...
-f, --reference=<referenceImagePath>
A reference image specifying the output size and resolution.
-o, --output=<outputFile>
Output file for transformed image
-q, --nThreads=<nThreads>
Number of threads.
-r, --output-resolution=<outputResolution>[,<outputResolution>...]
The resolution at which to write the output. Overrides reference image.
-s, --outputImageSize=<outputSize>
Size / field of view of image output in pixels. Comma separated, e.g.
"200,250,300". Overrides reference image.
-t, --transform=<transformFiles>
Transformation file.
transform2Dfield
-t <deformation field>?<inverse-options>?i
-o <output file>
Read about details of iterative inverses here.
A demonstration can be found here using this sample data.
Some common operations on swc neuron file are implemented by the script processNeuron:
Usage: <main class> [-h] [--set-radius=<radiusValue>] [-r=<radiusScaling>[,
<radiusScaling>...]] [-c=<coordinateScaling>[,
<coordinateScaling>...]]... [-o=<outputSkeletonPaths>]...
[-s=<skeletonPaths>]...
--set-radius=<radiusValue>
Set radius. The radius at every point will be set to this
value. This takes precedence over radius scaling.
-c=<coordinateScaling>[,<coordinateScaling>...]
Coordinate scaling
-h, --help Print this help message
-o=<outputSkeletonPaths>
Output skeletons. You can
-r=<radiusScaling>[,<radiusScaling>...]
Radius scaling
-s=<skeletonPaths> Input skeletons. You can provide multiple -s options
Scale x, y, and z coordinates by factors of 2, 3, and 4, respectively.
processNeuron -s GadMARCM-F000122_seg001_03.swc \
-o GadMARCM-F000122_seg001_03_scaledCoordinates.swc \
-c 2,3,4
Scale radii by a factor of 10
processNeuron -s GadMARCM-F000122_seg001_03.swc \
-o GadMARCM-F000122_seg001_03_scaledCoordinates.swc \
-r 10
Set the value of the radius to 1.0 everywhere
processNeuron -s GadMARCM-F000122_seg001_03.swc \
-o GadMARCM-F000122_seg001_03_scaledCoordinates.swc \
--set-radius 1.0