-
Notifications
You must be signed in to change notification settings - Fork 2.6k
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
[Project] Medical semantic seg dataset: Rite (#2680)
- Loading branch information
1 parent
5a9cfa9
commit d934d10
Showing
8 changed files
with
375 additions
and
0 deletions.
There are no files selected for viewing
135 changes: 135 additions & 0 deletions
135
projects/medical/2d_image/fundus_photography/rite/README.md
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,135 @@ | ||
| # Retinal Images vessel Tree Extraction (RITE) | ||
|
|
||
| ## Description | ||
|
|
||
| This project supports **`Retinal Images vessel Tree Extraction (RITE) `**, which can be downloaded from [here](https://opendatalab.com/RITE). | ||
|
|
||
| ### Dataset Overview | ||
|
|
||
| The RITE (Retinal Images vessel Tree Extraction) is a database that enables comparative studies on segmentation or classification of arteries and veins on retinal fundus images, which is established based on the public available DRIVE database (Digital Retinal Images for Vessel Extraction). RITE contains 40 sets of images, equally separated into a training subset and a test subset, the same as DRIVE. The two subsets are built from the corresponding two subsets in DRIVE. For each set, there is a fundus photograph, a vessel reference standard. The fundus photograph is inherited from DRIVE. For the training set, the vessel reference standard is a modified version of 1st_manual from DRIVE. For the test set, the vessel reference standard is 2nd_manual from DRIVE. | ||
|
|
||
| ### Statistic Information | ||
|
|
||
| | Dataset Name | Anatomical Region | Task Type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | | ||
| | ------------------------------------ | ----------------- | ------------ | ------------------ | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- | | ||
| | [Rite](https://opendatalab.com/RITE) | head_and_neck | segmentation | fundus_photography | 2 | 20/-/20 | yes/-/yes | 2013 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) | | ||
|
|
||
| | Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | | ||
| | :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | | ||
| | background | 20 | 91.61 | - | - | 20 | 91.58 | | ||
| | vessel | 20 | 8.39 | - | - | 20 | 8.42 | | ||
|
|
||
| Note: | ||
|
|
||
| - `Pct` means percentage of pixels in this category in all pixels. | ||
|
|
||
| ### Visualization | ||
|
|
||
| 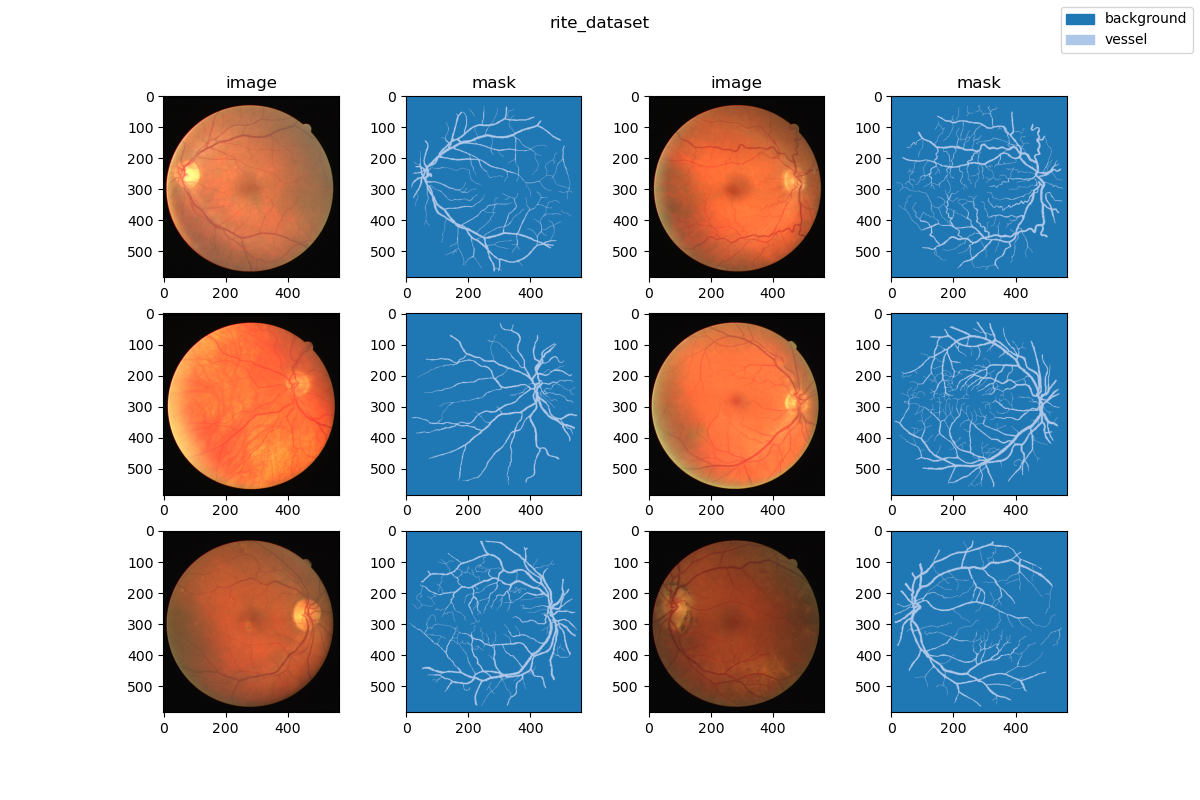 | ||
|
|
||
| ### Dataset Citation | ||
|
|
||
| ``` | ||
| @InProceedings{10.1007/978-3-642-40763-5_54, | ||
| author={Hu, Qiao and Abr{\`a}moff, Michael D. and Garvin, Mona K.}, | ||
| title={Automated Separation of Binary Overlapping Trees in Low-Contrast Color Retinal Images}, | ||
| booktitle={Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2013}, | ||
| year={2013}, | ||
| pages={436--443}, | ||
| } | ||
| ``` | ||
|
|
||
| ### Prerequisites | ||
|
|
||
| - Python v3.8 | ||
| - PyTorch v1.10.0 | ||
| - pillow(PIL) v9.3.0 9.3.0 | ||
| - scikit-learn(sklearn) v1.2.0 1.2.0 | ||
| - [MIM](https://github.com/open-mmlab/mim) v0.3.4 | ||
| - [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 | ||
| - [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher | ||
| - [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 | ||
|
|
||
| All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `rite/` root directory, run the following line to add the current directory to `PYTHONPATH`: | ||
|
|
||
| ```shell | ||
| export PYTHONPATH=`pwd`:$PYTHONPATH | ||
| ``` | ||
|
|
||
| ### Dataset Preparing | ||
|
|
||
| - download dataset from [here](https://opendatalab.com/RITE) and decompress data to path `'data/'`. | ||
| - run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. | ||
| - run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly. | ||
|
|
||
| ```none | ||
| mmsegmentation | ||
| ├── mmseg | ||
| ├── projects | ||
| │ ├── medical | ||
| │ │ ├── 2d_image | ||
| │ │ │ ├── fundus_photography | ||
| │ │ │ │ ├── rite | ||
| │ │ │ │ │ ├── configs | ||
| │ │ │ │ │ ├── datasets | ||
| │ │ │ │ │ ├── tools | ||
| │ │ │ │ │ ├── data | ||
| │ │ │ │ │ │ ├── train.txt | ||
| │ │ │ │ │ │ ├── val.txt | ||
| │ │ │ │ │ │ ├── images | ||
| │ │ │ │ │ │ │ ├── train | ||
| │ │ │ │ | │ │ │ ├── xxx.png | ||
| │ │ │ │ | │ │ │ ├── ... | ||
| │ │ │ │ | │ │ │ └── xxx.png | ||
| │ │ │ │ │ │ ├── masks | ||
| │ │ │ │ │ │ │ ├── train | ||
| │ │ │ │ | │ │ │ ├── xxx.png | ||
| │ │ │ │ | │ │ │ ├── ... | ||
| │ │ │ │ | │ │ │ └── xxx.png | ||
| ``` | ||
|
|
||
| ### Training commands | ||
|
|
||
| To train models on a single server with one GPU. (default) | ||
|
|
||
| ```shell | ||
| mim train mmseg ./configs/${CONFIG_FILE} | ||
| ``` | ||
|
|
||
| ### Testing commands | ||
|
|
||
| To test models on a single server with one GPU. (default) | ||
|
|
||
| ```shell | ||
| mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} | ||
| ``` | ||
|
|
||
| <!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) | ||
| You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> | ||
|
|
||
| ## Checklist | ||
|
|
||
| - [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. | ||
|
|
||
| - [x] Finish the code | ||
| - [x] Basic docstrings & proper citation | ||
| - [ ] Test-time correctness | ||
| - [x] A full README | ||
|
|
||
| - [ ] Milestone 2: Indicates a successful model implementation. | ||
|
|
||
| - [ ] Training-time correctness | ||
|
|
||
| - [ ] Milestone 3: Good to be a part of our core package! | ||
|
|
||
| - [ ] Type hints and docstrings | ||
| - [ ] Unit tests | ||
| - [ ] Code polishing | ||
| - [ ] Metafile.yml | ||
|
|
||
| - [ ] Move your modules into the core package following the codebase's file hierarchy structure. | ||
|
|
||
| - [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
17 changes: 17 additions & 0 deletions
17
...age/fundus_photography/rite/configs/fcn-unet-s5-d16_unet_1xb16-0.0001-20k_rite-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './rite_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.rite_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.0001) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
17 changes: 17 additions & 0 deletions
17
...mage/fundus_photography/rite/configs/fcn-unet-s5-d16_unet_1xb16-0.001-20k_rite-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './rite_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.rite_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.001) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
17 changes: 17 additions & 0 deletions
17
...image/fundus_photography/rite/configs/fcn-unet-s5-d16_unet_1xb16-0.01-20k_rite-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './rite_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.rite_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.01) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
18 changes: 18 additions & 0 deletions
18
...us_photography/rite/configs/fcn-unet-s5-d16_unet_1xb16-0.01lr-sigmoid-20k_rite-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './rite_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.rite_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.01) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict( | ||
| num_classes=2, loss_decode=dict(use_sigmoid=True), out_channels=1), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
42 changes: 42 additions & 0 deletions
42
projects/medical/2d_image/fundus_photography/rite/configs/rite_512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,42 @@ | ||
| dataset_type = 'RITEDataset' | ||
| data_root = 'data/' | ||
| img_scale = (512, 512) | ||
| train_pipeline = [ | ||
| dict(type='LoadImageFromFile'), | ||
| dict(type='LoadAnnotations'), | ||
| dict(type='Resize', scale=img_scale, keep_ratio=False), | ||
| dict(type='RandomFlip', prob=0.5), | ||
| dict(type='PhotoMetricDistortion'), | ||
| dict(type='PackSegInputs') | ||
| ] | ||
| test_pipeline = [ | ||
| dict(type='LoadImageFromFile'), | ||
| dict(type='Resize', scale=img_scale, keep_ratio=False), | ||
| dict(type='LoadAnnotations'), | ||
| dict(type='PackSegInputs') | ||
| ] | ||
| train_dataloader = dict( | ||
| batch_size=16, | ||
| num_workers=4, | ||
| persistent_workers=True, | ||
| sampler=dict(type='InfiniteSampler', shuffle=True), | ||
| dataset=dict( | ||
| type=dataset_type, | ||
| data_root=data_root, | ||
| ann_file='train.txt', | ||
| data_prefix=dict(img_path='images/', seg_map_path='masks/'), | ||
| pipeline=train_pipeline)) | ||
| val_dataloader = dict( | ||
| batch_size=1, | ||
| num_workers=4, | ||
| persistent_workers=True, | ||
| sampler=dict(type='DefaultSampler', shuffle=False), | ||
| dataset=dict( | ||
| type=dataset_type, | ||
| data_root=data_root, | ||
| ann_file='test.txt', | ||
| data_prefix=dict(img_path='images/', seg_map_path='masks/'), | ||
| pipeline=test_pipeline)) | ||
| test_dataloader = val_dataloader | ||
| val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice']) | ||
| test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice']) |
31 changes: 31 additions & 0 deletions
31
projects/medical/2d_image/fundus_photography/rite/datasets/rite_dataset.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,31 @@ | ||
| from mmseg.datasets import BaseSegDataset | ||
| from mmseg.registry import DATASETS | ||
|
|
||
|
|
||
| @DATASETS.register_module() | ||
| class RITEDataset(BaseSegDataset): | ||
| """RITEDataset dataset. | ||
| In segmentation map annotation for RITEDataset, | ||
| 0 stands for background, which is included in 2 categories. | ||
| ``reduce_zero_label`` is fixed to False. The ``img_suffix`` | ||
| is fixed to '.png' and ``seg_map_suffix`` is fixed to '.png'. | ||
| Args: | ||
| img_suffix (str): Suffix of images. Default: '.png' | ||
| seg_map_suffix (str): Suffix of segmentation maps. Default: '.png' | ||
| reduce_zero_label (bool): Whether to mark label zero as ignored. | ||
| Default to False. | ||
| """ | ||
| METAINFO = dict(classes=('background', 'vessel')) | ||
|
|
||
| def __init__(self, | ||
| img_suffix='.png', | ||
| seg_map_suffix='.png', | ||
| reduce_zero_label=False, | ||
| **kwargs) -> None: | ||
| super().__init__( | ||
| img_suffix=img_suffix, | ||
| seg_map_suffix=seg_map_suffix, | ||
| reduce_zero_label=reduce_zero_label, | ||
| **kwargs) |
98 changes: 98 additions & 0 deletions
98
projects/medical/2d_image/fundus_photography/rite/tools/prepare_dataset.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,98 @@ | ||
| import glob | ||
| import os | ||
|
|
||
| import numpy as np | ||
| from PIL import Image | ||
|
|
||
| root_path = 'data/' | ||
| img_suffix = '.tif' | ||
| seg_map_suffix = '.png' | ||
| save_img_suffix = '.png' | ||
| save_seg_map_suffix = '.png' | ||
| src_img_train_dir = os.path.join(root_path, 'AV_groundTruth/training/images/') | ||
| src_img_test_dir = os.path.join(root_path, 'AV_groundTruth/test/images/') | ||
| src_mask_train_dir = os.path.join(root_path, 'AV_groundTruth/training/vessel/') | ||
| src_mask_test_dir = os.path.join(root_path, 'AV_groundTruth/test/vessel/') | ||
|
|
||
| tgt_img_train_dir = os.path.join(root_path, 'images/train/') | ||
| tgt_mask_train_dir = os.path.join(root_path, 'masks/train/') | ||
| tgt_img_test_dir = os.path.join(root_path, 'images/test/') | ||
| tgt_mask_test_dir = os.path.join(root_path, 'masks/test/') | ||
| os.system('mkdir -p ' + tgt_img_train_dir) | ||
| os.system('mkdir -p ' + tgt_mask_train_dir) | ||
| os.system('mkdir -p ' + tgt_img_test_dir) | ||
| os.system('mkdir -p ' + tgt_mask_test_dir) | ||
|
|
||
|
|
||
| def filter_suffix_recursive(src_dir, suffix): | ||
| # filter out file names and paths in source directory | ||
| suffix = '.' + suffix if '.' not in suffix else suffix | ||
| file_paths = glob.glob( | ||
| os.path.join(src_dir, '**', '*' + suffix), recursive=True) | ||
| file_names = [_.split('/')[-1] for _ in file_paths] | ||
| return sorted(file_paths), sorted(file_names) | ||
|
|
||
|
|
||
| def convert_label(img, convert_dict): | ||
| arr = np.zeros_like(img, dtype=np.uint8) | ||
| for c, i in convert_dict.items(): | ||
| arr[img == c] = i | ||
| return arr | ||
|
|
||
|
|
||
| def convert_pics_into_pngs(src_dir, tgt_dir, suffix, convert='RGB'): | ||
| if not os.path.exists(tgt_dir): | ||
| os.makedirs(tgt_dir) | ||
|
|
||
| src_paths, src_names = filter_suffix_recursive(src_dir, suffix=suffix) | ||
| for i, (src_name, src_path) in enumerate(zip(src_names, src_paths)): | ||
| tgt_name = src_name.replace(suffix, save_img_suffix) | ||
| tgt_path = os.path.join(tgt_dir, tgt_name) | ||
| num = len(src_paths) | ||
| img = np.array(Image.open(src_path)) | ||
| if len(img.shape) == 2: | ||
| pil = Image.fromarray(img).convert(convert) | ||
| elif len(img.shape) == 3: | ||
| pil = Image.fromarray(img) | ||
| else: | ||
| raise ValueError('Input image not 2D/3D: ', img.shape) | ||
|
|
||
| pil.save(tgt_path) | ||
| print(f'processed {i+1}/{num}.') | ||
|
|
||
|
|
||
| def convert_label_pics_into_pngs(src_dir, | ||
| tgt_dir, | ||
| suffix, | ||
| convert_dict={ | ||
| 0: 0, | ||
| 255: 1 | ||
| }): | ||
| if not os.path.exists(tgt_dir): | ||
| os.makedirs(tgt_dir) | ||
|
|
||
| src_paths, src_names = filter_suffix_recursive(src_dir, suffix=suffix) | ||
| num = len(src_paths) | ||
| for i, (src_name, src_path) in enumerate(zip(src_names, src_paths)): | ||
| tgt_name = src_name.replace(suffix, save_seg_map_suffix) | ||
| tgt_path = os.path.join(tgt_dir, tgt_name) | ||
|
|
||
| img = np.array(Image.open(src_path)) | ||
| img = convert_label(img, convert_dict) | ||
| Image.fromarray(img).save(tgt_path) | ||
| print(f'processed {i+1}/{num}.') | ||
|
|
||
|
|
||
| if __name__ == '__main__': | ||
|
|
||
| convert_pics_into_pngs( | ||
| src_img_train_dir, tgt_img_train_dir, suffix=img_suffix) | ||
|
|
||
| convert_pics_into_pngs( | ||
| src_img_test_dir, tgt_img_test_dir, suffix=img_suffix) | ||
|
|
||
| convert_label_pics_into_pngs( | ||
| src_mask_train_dir, tgt_mask_train_dir, suffix=seg_map_suffix) | ||
|
|
||
| convert_label_pics_into_pngs( | ||
| src_mask_test_dir, tgt_mask_test_dir, suffix=seg_map_suffix) |