-
Notifications
You must be signed in to change notification settings - Fork 2.6k
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
[Project] Medical semantic seg dataset: Crass (#2690)
- Loading branch information
1 parent
c1de52a
commit c923f4d
Showing
8 changed files
with
369 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,144 @@ | ||
| # Chest Radiograph Anatomical Structure Segmentation (CRASS) | ||
|
|
||
| ## Description | ||
|
|
||
| This project supports **`Chest Radiograph Anatomical Structure Segmentation (CRASS) `**, which can be downloaded from [here](https://crass.grand-challenge.org/). | ||
|
|
||
| ### Dataset Overview | ||
|
|
||
| A set of consecutively obtained posterior-anterior chest radiograph were selected from a database containing images acquired at two sites in sub Saharan Africa with a high tuberculosis incidence. All subjects were 15 years or older. Images from digital chest radiography units were used (Delft Imaging Systems, The Netherlands) of varying resolutions, with a typical resolution of 1800--2000 pixels, the pixel size was 250 lm isotropic. From the total set of images, 225 were considered to be normal by an expert radiologist, while 333 of the images contained abnormalities. Of the abnormal images, 220 contained abnormalities in the upper area of the lung where the clavicle is located. The data was divided into a training and a test set. The training set consisted of 299 images, the test set of 249 images. | ||
| The current data is still incomplete and to be added later. | ||
|
|
||
| ### Information Statistics | ||
|
|
||
| | Dataset Name | Anatomical Region | Task Type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | | ||
| | ------------------------------------------- | ----------------- | ------------ | -------- | ------------ | --------------------- | ---------------------- | ------------ | ------------------------------------------------------------- | | ||
| | [crass](https://crass.grand-challenge.org/) | pulmonary | segmentation | x_ray | 2 | 299/-/234 | yes/-/no | 2021 | [CC0 1.0](https://creativecommons.org/publicdomain/zero/1.0/) | | ||
|
|
||
| | Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | | ||
| | :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | | ||
| | background | 299 | 98.38 | - | - | - | - | | ||
| | clavicles | 299 | 1.62 | - | - | - | - | | ||
|
|
||
| Note: | ||
|
|
||
| - `Pct` means percentage of pixels in this category in all pixels. | ||
|
|
||
| ### Visualization | ||
|
|
||
| 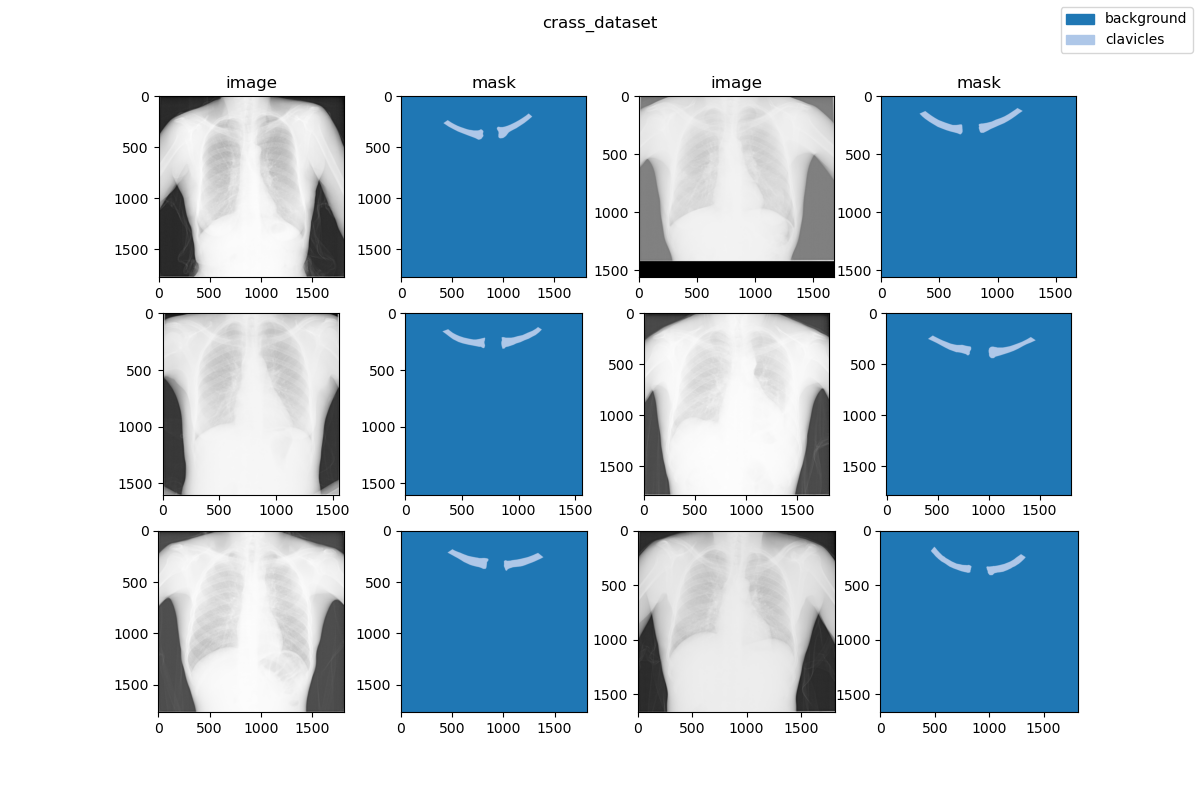 | ||
|
|
||
| ### Dataset Citation | ||
|
|
||
| ``` | ||
| @article{HOGEWEG20121490, | ||
| title={Clavicle segmentation in chest radiographs}, | ||
| journal={Medical Image Analysis}, | ||
| volume={16}, | ||
| number={8}, | ||
| pages={1490-1502}, | ||
| year={2012} | ||
| } | ||
| ``` | ||
|
|
||
| ### Prerequisites | ||
|
|
||
| - Python v3.8 | ||
| - PyTorch v1.10.0 | ||
| - pillow(PIL) v9.3.0 | ||
| - scikit-learn(sklearn) v1.2.0 | ||
| - [MIM](https://github.com/open-mmlab/mim) v0.3.4 | ||
| - [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 | ||
| - [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher | ||
| - [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 | ||
|
|
||
| All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `crass/` root directory, run the following line to add the current directory to `PYTHONPATH`: | ||
|
|
||
| ```shell | ||
| export PYTHONPATH=`pwd`:$PYTHONPATH | ||
| ``` | ||
|
|
||
| ### Dataset Preparing | ||
|
|
||
| - download dataset from [here](https://crass.grand-challenge.org/) and decompress data to path `'data/'`. | ||
| - run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. | ||
| - run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly. | ||
|
|
||
| ```none | ||
| mmsegmentation | ||
| ├── mmseg | ||
| ├── projects | ||
| │ ├── medical | ||
| │ │ ├── 2d_image | ||
| │ │ │ ├── x_ray | ||
| │ │ │ │ ├── crass | ||
| │ │ │ │ │ ├── configs | ||
| │ │ │ │ │ ├── datasets | ||
| │ │ │ │ │ ├── tools | ||
| │ │ │ │ │ ├── data | ||
| │ │ │ │ │ │ ├── train.txt | ||
| │ │ │ │ │ │ ├── val.txt | ||
| │ │ │ │ │ │ ├── images | ||
| │ │ │ │ │ │ │ ├── train | ||
| │ │ │ │ | │ │ │ ├── xxx.png | ||
| │ │ │ │ | │ │ │ ├── ... | ||
| │ │ │ │ | │ │ │ └── xxx.png | ||
| │ │ │ │ │ │ ├── masks | ||
| │ │ │ │ │ │ │ ├── train | ||
| │ │ │ │ | │ │ │ ├── xxx.png | ||
| │ │ │ │ | │ │ │ ├── ... | ||
| │ │ │ │ | │ │ │ └── xxx.png | ||
| ``` | ||
|
|
||
| ### Divided Dataset Information | ||
|
|
||
| ***Note: The table information below is divided by ourselves.*** | ||
|
|
||
| | Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | | ||
| | :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | | ||
| | background | 227 | 98.38 | 57 | 98.39 | - | - | | ||
| | clavicles | 227 | 1.62 | 57 | 1.61 | - | - | | ||
|
|
||
| ### Training commands | ||
|
|
||
| To train models on a single server with one GPU. (default) | ||
|
|
||
| ```shell | ||
| mim train mmseg ./configs/${CONFIG_FILE} | ||
| ``` | ||
|
|
||
| ### Testing commands | ||
|
|
||
| To test models on a single server with one GPU. (default) | ||
|
|
||
| ```shell | ||
| mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} | ||
| ``` | ||
|
|
||
| <!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) | ||
| You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> | ||
|
|
||
| ## Checklist | ||
|
|
||
| - [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. | ||
|
|
||
| - [x] Finish the code | ||
| - [x] Basic docstrings & proper citation | ||
| - [ ] Test-time correctness | ||
| - [x] A full README | ||
|
|
||
| - [x] Milestone 2: Indicates a successful model implementation. | ||
|
|
||
| - [x] Training-time correctness | ||
|
|
||
| - [ ] Milestone 3: Good to be a part of our core package! | ||
|
|
||
| - [ ] Type hints and docstrings | ||
| - [ ] Unit tests | ||
| - [ ] Code polishing | ||
| - [ ] Metafile.yml | ||
|
|
||
| - [ ] Move your modules into the core package following the codebase's file hierarchy structure. | ||
|
|
||
| - [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
42 changes: 42 additions & 0 deletions
42
projects/medical/2d_image/x_ray/crass/configs/crass_512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,42 @@ | ||
| dataset_type = 'CRASSDataset' | ||
| data_root = 'data/' | ||
| img_scale = (512, 512) | ||
| train_pipeline = [ | ||
| dict(type='LoadImageFromFile'), | ||
| dict(type='LoadAnnotations'), | ||
| dict(type='Resize', scale=img_scale, keep_ratio=False), | ||
| dict(type='RandomFlip', prob=0.5), | ||
| dict(type='PhotoMetricDistortion'), | ||
| dict(type='PackSegInputs') | ||
| ] | ||
| test_pipeline = [ | ||
| dict(type='LoadImageFromFile'), | ||
| dict(type='Resize', scale=img_scale, keep_ratio=False), | ||
| dict(type='LoadAnnotations'), | ||
| dict(type='PackSegInputs') | ||
| ] | ||
| train_dataloader = dict( | ||
| batch_size=16, | ||
| num_workers=4, | ||
| persistent_workers=True, | ||
| sampler=dict(type='InfiniteSampler', shuffle=True), | ||
| dataset=dict( | ||
| type=dataset_type, | ||
| data_root=data_root, | ||
| ann_file='train.txt', | ||
| data_prefix=dict(img_path='images/', seg_map_path='masks/'), | ||
| pipeline=train_pipeline)) | ||
| val_dataloader = dict( | ||
| batch_size=1, | ||
| num_workers=4, | ||
| persistent_workers=True, | ||
| sampler=dict(type='DefaultSampler', shuffle=False), | ||
| dataset=dict( | ||
| type=dataset_type, | ||
| data_root=data_root, | ||
| ann_file='tval.txt', | ||
| data_prefix=dict(img_path='images/', seg_map_path='masks/'), | ||
| pipeline=test_pipeline)) | ||
| test_dataloader = val_dataloader | ||
| val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice']) | ||
| test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice']) |
17 changes: 17 additions & 0 deletions
17
...dical/2d_image/x_ray/crass/configs/fcn-unet-s5-d16_unet_1xb16-0.0001-20k_crass-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './crass_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.crass_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.0001) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
17 changes: 17 additions & 0 deletions
17
...edical/2d_image/x_ray/crass/configs/fcn-unet-s5-d16_unet_1xb16-0.001-20k_crass-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './crass_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.crass_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.001) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
17 changes: 17 additions & 0 deletions
17
...medical/2d_image/x_ray/crass/configs/fcn-unet-s5-d16_unet_1xb16-0.01-20k_crass-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './crass_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.crass_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.01) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
18 changes: 18 additions & 0 deletions
18
..._image/x_ray/crass/configs/fcn-unet-s5-d16_unet_1xb16-lr0.01-sigmoid-20k_crass-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', './crass_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.crass_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.01) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict( | ||
| num_classes=2, loss_decode=dict(use_sigmoid=True), out_channels=1), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
30 changes: 30 additions & 0 deletions
30
projects/medical/2d_image/x_ray/crass/datasets/crass_dataset.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,30 @@ | ||
| from mmseg.datasets import BaseSegDataset | ||
| from mmseg.registry import DATASETS | ||
|
|
||
|
|
||
| @DATASETS.register_module() | ||
| class CRASSDataset(BaseSegDataset): | ||
| """CRASSDataset dataset. | ||
| In segmentation map annotation for CRASSDataset, 0 stands for background, | ||
| which is included in 2 categories. ``reduce_zero_label`` is fixed to | ||
| False. The ``img_suffix`` is fixed to '.png' and ``seg_map_suffix`` is | ||
| fixed to '.png'. | ||
| Args: | ||
| img_suffix (str): Suffix of images. Default: '.png' | ||
| seg_map_suffix (str): Suffix of segmentation maps. Default: '.png' | ||
| reduce_zero_label (bool): Whether to mark label zero as ignored. | ||
| Default to False.. | ||
| """ | ||
| METAINFO = dict(classes=('background', 'clavicles')) | ||
|
|
||
| def __init__(self, | ||
| img_suffix='.png', | ||
| seg_map_suffix='.png', | ||
| reduce_zero_label=False, | ||
| **kwargs) -> None: | ||
| super().__init__( | ||
| img_suffix=img_suffix, | ||
| seg_map_suffix=seg_map_suffix, | ||
| reduce_zero_label=reduce_zero_label, | ||
| **kwargs) |
84 changes: 84 additions & 0 deletions
84
projects/medical/2d_image/x_ray/crass/tools/prepare_dataset.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,84 @@ | ||
| import glob | ||
| import os | ||
|
|
||
| import cv2 | ||
| import SimpleITK as sitk | ||
| from PIL import Image | ||
|
|
||
| root_path = 'data/' | ||
| img_suffix = '.tif' | ||
| seg_map_suffix = '.png' | ||
| save_img_suffix = '.png' | ||
| save_seg_map_suffix = '.png' | ||
|
|
||
| src_img_train_dir = os.path.join(root_path, 'CRASS/data_train') | ||
| src_mask_train_dir = os.path.join(root_path, 'CRASS/mask_mhd') | ||
| src_img_test_dir = os.path.join(root_path, 'CRASS/data_test') | ||
|
|
||
| tgt_img_train_dir = os.path.join(root_path, 'images/train/') | ||
| tgt_mask_train_dir = os.path.join(root_path, 'masks/train/') | ||
| tgt_img_test_dir = os.path.join(root_path, 'images/test/') | ||
| os.system('mkdir -p ' + tgt_img_train_dir) | ||
| os.system('mkdir -p ' + tgt_mask_train_dir) | ||
| os.system('mkdir -p ' + tgt_img_test_dir) | ||
|
|
||
|
|
||
| def filter_suffix_recursive(src_dir, suffix): | ||
| suffix = '.' + suffix if '.' not in suffix else suffix | ||
| file_paths = glob( | ||
| os.path.join(src_dir, '**', '*' + suffix), recursive=True) | ||
| file_names = [_.split('/')[-1] for _ in file_paths] | ||
| return sorted(file_paths), sorted(file_names) | ||
|
|
||
|
|
||
| def read_single_array_from_med(path): | ||
| return sitk.GetArrayFromImage(sitk.ReadImage(path)).squeeze() | ||
|
|
||
|
|
||
| def convert_meds_into_pngs(src_dir, | ||
| tgt_dir, | ||
| suffix='.dcm', | ||
| norm_min=0, | ||
| norm_max=255, | ||
| convert='RGB'): | ||
| if not os.path.exists(tgt_dir): | ||
| os.makedirs(tgt_dir) | ||
|
|
||
| src_paths, src_names = filter_suffix_recursive(src_dir, suffix=suffix) | ||
| num = len(src_paths) | ||
| for i, (src_name, src_path) in enumerate(zip(src_names, src_paths)): | ||
| tgt_name = src_name.replace(suffix, '.png') | ||
| tgt_path = os.path.join(tgt_dir, tgt_name) | ||
|
|
||
| img = read_single_array_from_med(src_path) | ||
| if norm_min is not None and norm_max is not None: | ||
| img = cv2.normalize(img, None, norm_min, norm_max, cv2.NORM_MINMAX, | ||
| cv2.CV_8U) | ||
| pil = Image.fromarray(img).convert(convert) | ||
| pil.save(tgt_path) | ||
| print(f'processed {i+1}/{num}.') | ||
|
|
||
|
|
||
| convert_meds_into_pngs( | ||
| src_img_train_dir, | ||
| tgt_img_train_dir, | ||
| suffix='.mhd', | ||
| norm_min=0, | ||
| norm_max=255, | ||
| convert='RGB') | ||
|
|
||
| convert_meds_into_pngs( | ||
| src_img_test_dir, | ||
| tgt_img_test_dir, | ||
| suffix='.mhd', | ||
| norm_min=0, | ||
| norm_max=255, | ||
| convert='RGB') | ||
|
|
||
| convert_meds_into_pngs( | ||
| src_mask_train_dir, | ||
| tgt_mask_train_dir, | ||
| suffix='.mhd', | ||
| norm_min=0, | ||
| norm_max=1, | ||
| convert='L') |