(overview)=
Below we will first outline what you need to get started, the different ways you can use DeepLabCut, and then the full workflow. Note, we highly recommend you also read and follow our Nature Protocols paper, which is (still) fully relevant to standard DeepLabCut.
-
Decide on your needs: there are two main modes, standard DeepLabCut or multi-animal DeepLabCut. We highly recommend carefully considering which one is best for your needs. For example, a white mouse + black mouse would call for standard, while two black mice would use multi-animal. Important Information on how to use DLC in different scenarios (single vs multi animal) Then pick:
Additional Learning Resources:
- TUTORIALS: video tutorials that demonstrate various aspects of using the code base.
- HOW-TO-GUIDES: step-by-step user guidelines for using DeepLabCut on your own datasets (see below)
- EXPLANATIONS: resources on understanding how DeepLabCut works
- REFERENCES: read the science behind DeepLabCut
Getting Started: a video tutorial on navigating the documentation!
-
a set of videos that span the types of behaviors you want to track. Having 10 videos that include different backgrounds, different individuals, and different postures is MUCH better than 1 or 2 videos of 1 or 2 different individuals (i.e. 10-20 frames from each of 10 videos is much better than 50-100 frames from 2 videos).
-
minimally, a computer w/a CPU. If you want to use DeepLabCut on your own computer for many experiments, then you should get an NVIDIA GPU. See technical specs here. You can also use cloud computing resources, including COLAB (see how).
-
no specific cameras/videos are required; color, monochrome, etc., is all fine. If you can see what you want to measure, then this will work for you (given enough labeled data).
-
no specific computer is required (but see recommendations above), our software works on Linux, Windows, and MacOS, although we recommend Ubuntu.
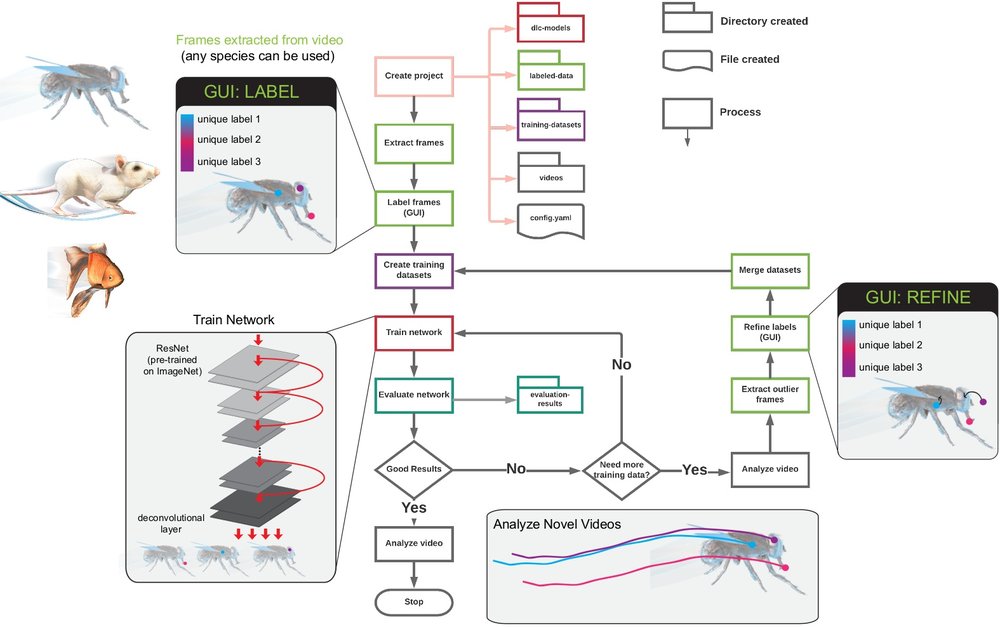
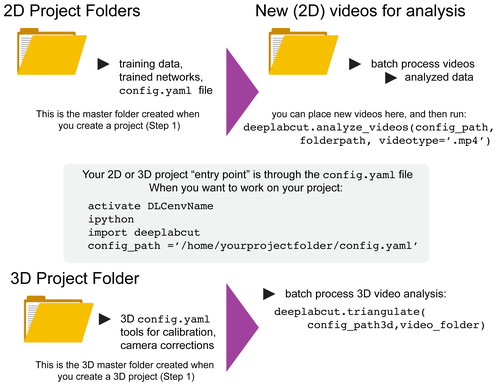
DeepLabCut is a software package for markerless pose estimation of animals performing various tasks. The software can manage multiple projects for various tasks. Each project is identified by the name of the project (e.g. TheBehavior), name of the experimenter (e.g. YourName), as well as the date at creation. This project folder holds a config.yaml (a text document) file containing various (project) parameters as well as links the data of the project.
This page contains a list of the essential functions of DeepLabCut as well as demos. There are many optional parameters with each described function, which you can find here. For additional assistance, you can use the help function to better understand what each function does.
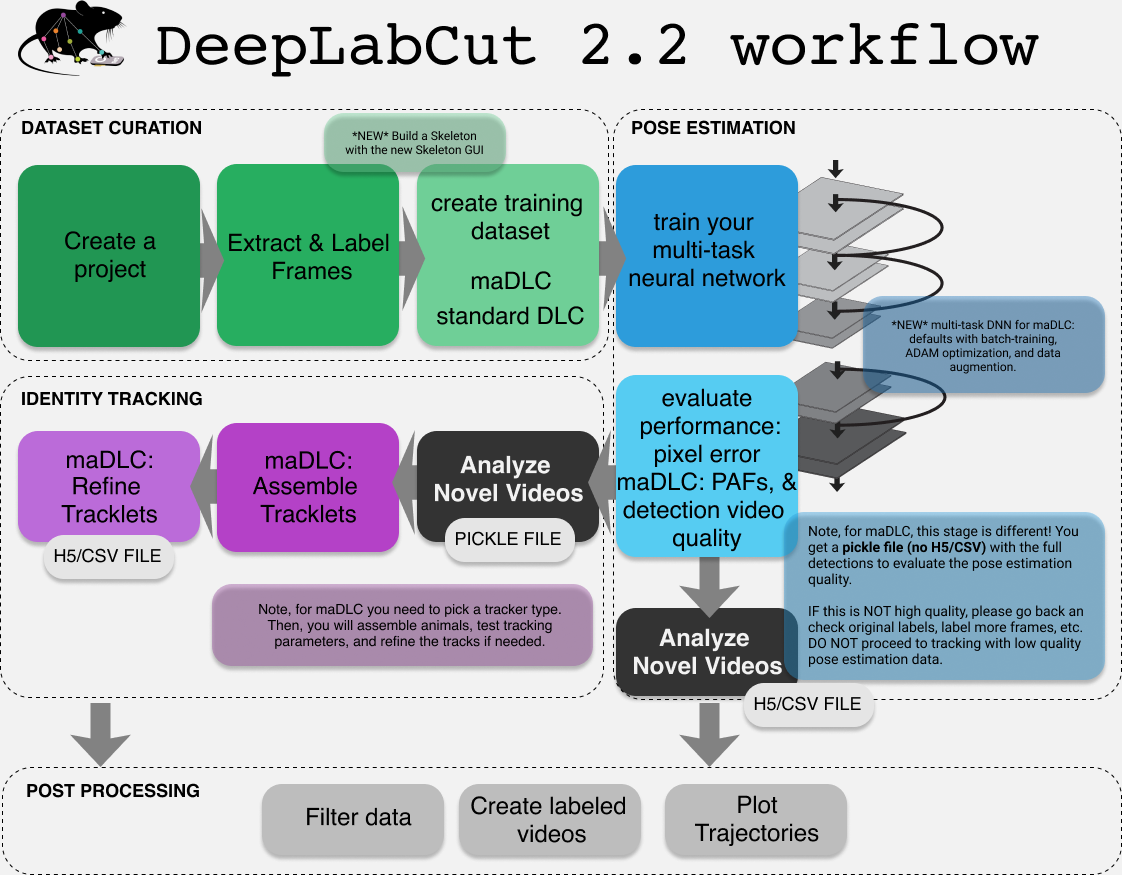
NOTE: There is a highly similar workflow for 2.2+ (and your 2.X projects are still fully compatible with this format!).
** DLC 2.2:** as of 2.2 we support "multi-animal projects," but these new features can also be used on single animals too (details below). The workflow is highly similar, but with a few key additional steps. Please carefully review the functions below for more details. You can search/look for maDeepLabCut for specific steps that are changed, or see this more comprehensive guide here
You can have as many projects on your computer as you wish. You can have DeepLabCut installed in an environment and always exit and return to this environment to run the code. You just need to point to the correct config.yaml file to jump back in! The documentation below will take you through the individual steps.
(important-info-regd-usage)=
We recommend first using DeepLabCut for a single animal scenario to understand the workflow - even if it's just our demo data. Multi-animal tracking is more complex - i.e. it has several decisions the user needs to make. Then, when you are ready you can jump into 2.2...
We highly recommend using 2.2 first in the Project Manager GUI (Option 3). This will allow you to get used to the additional steps by being walked through the process. Then, you can always use all the functions in your favorite IDE, notebooks, etc.
-
I have single animal videos:
- quick start: when you
create_new_project(and leave the default flag to False inmultianimal=False). This is the typical work path for many of you.
- quick start: when you
-
I have single animal videos, but I want to use the updated network capabilities introduced for multi-animal projects:
- quick start: when you
create_new_projectjust set the flagmultianimal=True. This enables you to use maDLC features even though you have only one animal. To note, this is rarely required for single animal projects, and not the recommended path. Some tips for when you might want to use this: this is good for say, a hand or a mouse if you feel the "skeleton" during training would increase performance. DON'T do this for things that could be identified an individual objects. i.e., don't do whisker 1, whisker 2, whisker 3 as 3 individuals. Each whisker always has a specific spatial location, and by calling them individuals you will do WORSE than in single animal mode.
- quick start: when you
- I have multiple identical-looking animals in my videos:
- quick start: when you
create_new_projectset the flagmultianimal=True. If you can't tell them apart, you can assign the "individual" ID to any animal in each frame. See this labeling w/2.2 demo video
- quick start: when you
- I have multiple animals, but I can tell them apart, in my videos and want to use DLC2.2:
- quick start: when you
create_new_projectset the flagmultianimal=True. And always label the "individual" ID name the same; i.e. if you have mouse1 and mouse2 but mouse2 always has a miniscope, in every frame label mouse2 consistently. See this labeling w/2.2 demo video. Then, you MUST put the following in the config.yaml file:identity: true
- quick start: when you
VIDEO TUTORIAL AVAILABLE! - ALSO, if you can tell them apart, label animals them consistently!
- I have a pre-2.2 single animal project, but I want to use 2.2:
Please read this convert 2 maDLC guide
Great - now that you get the overall workflow let's jump in! Here, you have several options.
Option 1 DEMOs: for a quick introduction to DLC on our data.
Option 2 Standalone GUI: is the perfect place for beginners who want to start using DeepLabCut on your own data.
Option 3 In the terminal: is best for more advanced users, as with the terminal interface you get the most versatility and options.
(using-demo-notebooks)=
We provide Jupyter and COLAB notebooks for using DeepLabCut on both a pre-labeled dataset, and on the end user’s own dataset. See all the demo's here! Please note that GUIs are not easily supported in Jupyter in MacOS, as you need a framework build of python. While it's possible to launch them with a few tweaks, we recommend using the Project Manager GUI or terminal, so please follow the instructions below.
(using-project-manager-gui)=
Start iPython, or if you are using MacOS, you must use pythonw vs. typing ipython or python, but otherwise it's the same.
If you are using DeepLabCut on the cloud, you cannot use the GUIs and you need to first set DLClight=True. If you use Windows, please always open the terminal with administrator privileges. Please read more here, and in our Nature Protocols paper here. And, see our troubleshooting wiki.
Simply open the terminal and type:
python -m deeplabcutThat's it! Follow the GUI for details
(using-the-terminal)=
Please decide with mode you want to use DeepLabCut, and follow one of the following: