SOVABIDS: A python package for the automatic conversion of MEG/EEG datasets to the BIDS standard, with a focus on making the most out of metadata
BIDS is a standard for neuroimaging datasets that helps with data sharing and reusability; as a result it has been widely adopted by the community. Although developed for MRI originally, it has gained popularity within the EEG and MEG realms. Converting raw data to the BIDS standard is not difficult but requires a lot of time if done by hand. Currently software like mne-bids and Biscuit are available to assist and facilitate this conversion but there is still no automated way to produce a valid BIDS dataset for the EEG and MEG use-cases. Mostly what is missing is metadata. Here a python package is proposed to be able to infer the missing information from files accompanying the EEG and MEG files and/or a single bids-conversion example provided by the user. The idea of having simple human-readable configuration files is also explored since this would help the sharing of common conversion parameters within similar communities. If this proposal was successfully implemented then batch conversion of BIDS datasets in the EEG and MEG cases would be realized. Moreover, since the design is constrained to simple configuration files, the software could potentially expand BIDS adoption to people not experienced with scripting. It is hoped that this package becomes the backend of an hypothetical web application that assists the conversion of MEG-EEG datasets to BIDS.
Data is a fundamental asset for science, sadly it is usually stored in such a way that only the original researcher knows how to make use of it; the neuroimaging community is no stranger to this phenomenon. As the accumulation and sharing of data becomes more important following the data-intensive needs of neuroimaging, it has become critical the existence of an unified way to store data among researchers. One of the solutions to this is the BIDS standard for neuroimaging data, originally developed for MRI. As the BIDS adoption increases, it expands its range of application; in particular the MEG and EEG extensions are becoming more popular in their respective communities.
In essence, BIDS is a specification that sets how data is saved on the storage medium: its folder structure, the filenames, the metadata format, etc. The BIDS standard is simple to understand but doing the conversion from a raw source may demand a lot of time. Luckily semi-automatic and automatic converters have been developed to address this, although mainly for MRI. As of now for MEG and EEG conversion some software packages have been developed: MNE-BIDS (working mainly by python scripts), Biscuit (GUI-based converter done in python), some plugins for EEGLAB, FieldTrip and SPM. and some manufacturer specific converters (BrainVision, for example). Most of these softwares will infer the technical characteristics to populate the BIDS dataset but will require the user to input the non-technical metadata (in example subjects ids, demographic characteristics,etc) through either scripting or GUI interfaces. The previous limitation makes the process of the -to BIDS- conversion complex and not completely automatic. In a lot of cases the information missing is available through metadata files, only needing a way for them to be read and incorporated into the already extracted technical information. In general the user ends up inputting a lot of information by hand or he needs to do scripts particular for his use-case. Since a lot of the MEG and EEG researchers don’t have a programming background this task will end up being manual. Moreover, the path containing the files usually encodes study design information, which is not usually used to improve the automation. MNE-BIDS for example will require inputting the subject,session and run through a python script, BISCUITS will require the user to set some of these parameters manually through a GUI and so on.
Another improvement that could be made for automatic “MEG/EEG datasets to BIDS” conversion is to have a file that encodes the conversion parameters, so that they can be easily shared within a community with the same storage pattern. In addition, some individual MEG/EEG records may have particular characteristics not shared between all the records, thus requiring another file that characterizes the conversion for a particular case. The two files mentioned previously should be simple and intuitive enough so that people not experienced with scripting and programming can make use of the software. Similarly, the learning curve of the software should be low enough for people to adopt it.
Motivated by the previous discussion, the proposed solution mainly addresses the following questions:
- How can we infer as much as we can from the metadata files available and from a single bids conversion example provided by the user?.
- How can we encode the information inferred previously in a simple and intuitive human-readable way?
- How can we encode in a file the “to-bids” mapping that setups the conversion of a single record ?
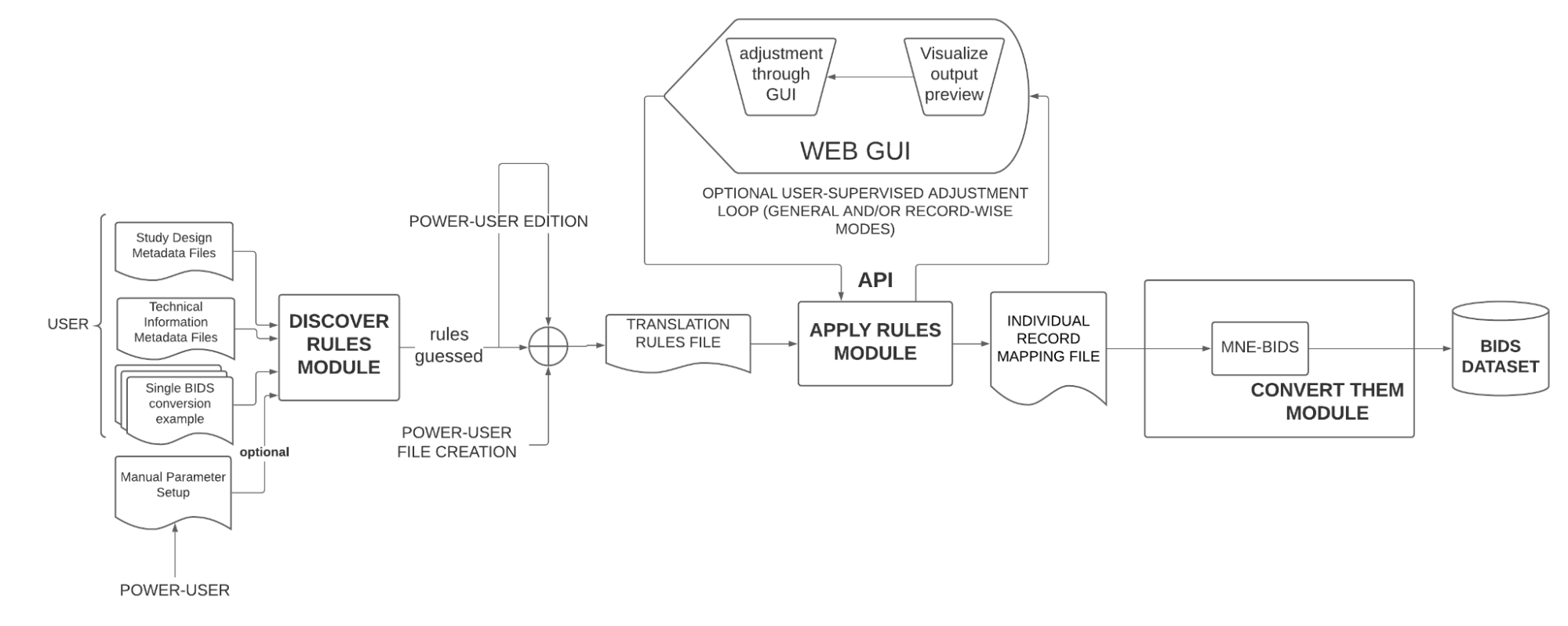
The following schematic illustrates the overall design of the proposed software:
The software is designed to work with EEG and MEG files, but because of the time constraints of the program, the GSoC project will focus on the conversion of EEG files from (at least) two different EEG system vendors. Moreover, the GUI of the “user-supervised adjustment loop”, although designed to be in the final version, won’t be developed during GSoC because of the same reason.
The three main modules illustrated previously are expanded accordingly:
This module is in charge of obtaining a “Translation Rules File” that describes the general rules for the conversion of the MEG/EEG dataset. For that it will infer characteristics from two sources:
- A single example of a successful bids conversion provided by the user.
- Metadata files available.
From the structural point of view, the metadata files contemplated here are:
- Manufacturer-Specific
- Tabular Files
- Dictionary Files
Internally, this module is made out of functions that will apply different heuristics to get the information of interest.
From the functional point of view two types of metadata arise:
- Study Design Metadata
- Technical Metadata of the Acquisition
Researchers are usually accustomed to the study design details, whereas the technical metadata may be known by a technical user. It is possible that either of these two users would want to set directly some parameters rather than them to be guessed. Because of this, the module should have a way to set directly some details rather than guess them.
This module interprets the “Translation Rules” file and outputs a “Individual Record Mapping” file. This is mainly applying the rules of the “Translation Rules File” to a single record.
Power-users may produce the “Translation Rules” file on their own if they wish to do so; thus skipping the “Discover Rules” module. This would be possible since the file is designed to be simple and intuitive syntax.
Since the “guessed” rules from the previous module are not necessarily correct in all cases, an user-supervised adjustment loop powered by a GUI is included in the design of the software. This GUI should show the user the hypothetical results of the conversion and offer a way to change the parameters so that the user gets the expected results. The changes should be applied on the “Translation Rule” if the change is a general one or on the “Individual Record Mapping” for record-wise changes.
The “Apply Rules” Module should plug into the mentioned GUI through an API so as to not limit the visualization possibilities of the package. That is, during this GSoC, an effort will be made to implement the necessary API to achieve this rather than on a GUI.
This module grabs the “Individual Record Mapping” file (or a collection of them) and performs the conversion using the mne-bids package. A way to override the inferences done by the mne-bids package module should be applied if the user specifies to do so.
Python package that contains the following modules:
- “Discover Rules” Module.
- “Apply Rules” Module.
- “Convert Them” Module.
- “Translation Rules” File Schema
- “Individual Record Mapping” File Schema
- Use-case examples illustrating the conversion process of files from 2 different EEG System Vendors
- Documentation.
If this software was successfully developed it would contribute to the adoption of the BIDS standard in EEG and MEG communities that don’t have a scripting/programming background. It would allow the automatic conversion of EEG/MEG datasets on servers since the conversion is controlled by configuration files. These same configurations files open the possibility for exchange of conversion mappings between communities that have a similar storage strategy.
I have some experience working with EEG data and how it is handled by BIDS. This experience was mostly acquired through an internship in a research group of the University Of Antioquia called “Neuropsychology and Behaviour Group” (GRUNECO). There I learned mostly resting EEG processing. Since the research group was (and is) trying to standardize their datasets I learned about the BIDS standard for EEG; coincidentally, I worked with a partner in this group to make a proof-of-concept software that does the conversion of a raw dataset to the BIDS standard. Although this software was a rudimentary initial design, it tackled problems similar to those explored in this proposal. I also have experience with python and in particular I contributed to some bug fixing and refactoring in the open source package “pyprep”. Through these last efforts I acquired some knowledge about the mne package workings. As a result of these experiences I have knowledge of the python language, the EEG files, the BIDS ecosystem and the mne package library; all of these are essential for the execution of this project.
Apart from the previous, I also collaborated with my mentor Oren Civier to build this proposal: starting from the initial conversations through the NeuroStars forum up to using google docs to collaborate.
Note: During the course of GSoC I will be attending my university classes.
This timeline provides a rough guideline of how the project will be done.
Exploration of common EEG metadata formats.
Learning of the BIDS specification for EEG.
Familiarisation with the mne-bids package to produce BIDS datasets.
Design of the “Translation Rules” file schema that defines the conversion process of the dataset from a general point of view.
Deliverables:
- “Translation Rules” File Schema
Design of the “Individual Record Mapping” file schema and development of the “Apply Rules” module that outputs mapping files for each record.
Deliverables:
- “Individual Record Mapping” File Schema
- “Apply Rules” Module
Development of the “Discover Rules” module with functions for each metadata file format explored and for the analysis of a single bids-conversion example provided by the user.
Deliverables:
- “Discover Rules” Module
Development of a module in charge of obtaining, through the mapping file, the specific parameters to be passed to the mne-bids package. This will store the data in the structure specified by the BIDS standard.
Deliverables:
- “Convert Records” Module
Testing and correction of possible errors found and development conversion examples.
Deliverables:
- Examples illustrating the conversion process of files from 2 different EEG System Vendors
Documentation of the software.
Deliverables:
- Documentation